Lab: DNA exercise
DNA exercise
# vector ... etc.
my_dna <- "AACGAATGAGTAAATGAGTAAATGAAGGAATGATTATTCCTTGCTTTAGAACTTCTGGAATTAGAGGACAATATTAATAATACCATCGCACAGTGTTTCTTTGTTGTTAATGCTACAACATACAAAGAGGAAGCATGCAG"
my_dna## [1] "AACGAATGAGTAAATGAGTAAATGAAGGAATGATTATTCCTTGCTTTAGAACTTCTGGAATTAGAGGACAATATTAATAATACCATCGCACAGTGTTTCTTTGTTGTTAATGCTACAACATACAAAGAGGAAGCATGCAG"## [1] 1## [1] "character"## chr "AACGAATGAGTAAATGAGTAAATGAAGGAATGATTATTCCTTGCTTTAGAACTTCTGGAATTAGAGGACAATATTAATAATACCATCGCACAGTGTTTCTTTGTTGTTAAT"| __truncated__## [1] 140## [1] 1## [1] "list"## [1] 140## chr [1:140] "A" "A" "C" "G" "A" "A" "T" "G" "A" "G" "T" "A" "A" "A" "T" "G" "A" "G" "T" "A" ...## [1] 140## [1] "A" "C" "G" "T"## [1] TRUE TRUE FALSE FALSE TRUE TRUE FALSE FALSE TRUE FALSE FALSE TRUE TRUE TRUE FALSE
## [16] FALSE TRUE FALSE FALSE TRUE TRUE TRUE FALSE FALSE TRUE TRUE FALSE FALSE TRUE TRUE
## [31] FALSE FALSE TRUE FALSE FALSE TRUE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
## [46] FALSE FALSE TRUE FALSE TRUE TRUE FALSE FALSE FALSE FALSE FALSE FALSE FALSE TRUE TRUE
## [61] FALSE FALSE TRUE FALSE TRUE FALSE FALSE TRUE FALSE TRUE TRUE FALSE TRUE FALSE FALSE
## [76] TRUE TRUE FALSE TRUE TRUE FALSE TRUE FALSE FALSE TRUE FALSE FALSE FALSE FALSE TRUE
## [91] FALSE TRUE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
## [106] FALSE FALSE FALSE TRUE TRUE FALSE FALSE FALSE FALSE TRUE FALSE TRUE TRUE FALSE TRUE
## [121] FALSE TRUE FALSE TRUE TRUE TRUE FALSE TRUE FALSE FALSE TRUE TRUE FALSE FALSE TRUE
## [136] FALSE FALSE FALSE TRUE FALSE## [1] 52Frequency distribution
A gene consists of a sequence of nucleotides {A, C, G, T}.The number of each nucleotide can be displayed in a frequency table. This will be illustrated by the Zyxin gene which plays an important role in cell adhesion (Golub et al., 1999). The accession number (X94991.1) of oneof its variants can be found via an NCBI UniGene search. The code below illustrates how to install the package ape, to load it, to read gene ”X94991.1”of the species homo sapiens from GenBank, and to make a frequency table of the four nucleotides
# install.packages(c("ape"), repo="http://cran.r-project.org", dep=TRUE)
library(ape)
gene <- read.GenBank(c("X94991.1"), as.character=TRUE)
table(gene)## gene
## a c g t
## 410 789 573 394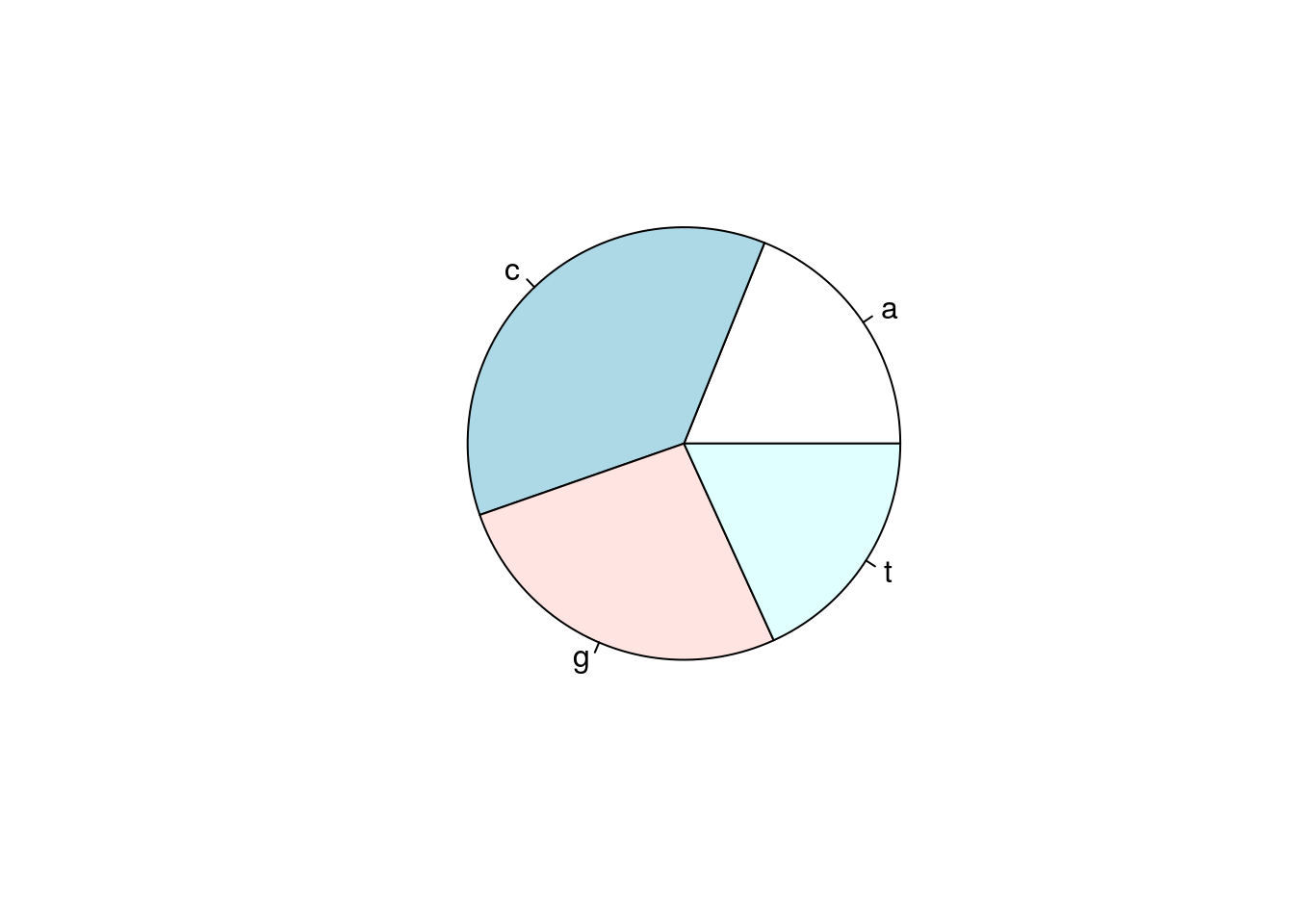
LS0tCnRpdGxlOiAiTGFiOiBETkEgZXhlcmNpc2UiCnBhZ2V0aXRsZTogIkxhYjogRE5BIGV4ZXJjaXNlIgotLS0KCmBgYHtyIHNldHVwLCBpbmNsdWRlPUZBTFNFfQpsaWJyYXJ5KGtuaXRyKQprbml0cjo6b3B0c19jaHVuayRzZXQoZWNobyA9IFRSVUUpCmBgYAoKIyMgRE5BIGV4ZXJjaXNlCmBgYHtyIGRuYV9leDIsIGluY2x1ZGUgPSBULCBlY2hvPVQsIGV2YWw9VH0KIyB2ZWN0b3IgLi4uIGV0Yy4KbXlfZG5hIDwtICJBQUNHQUFUR0FHVEFBQVRHQUdUQUFBVEdBQUdHQUFUR0FUVEFUVENDVFRHQ1RUVEFHQUFDVFRDVEdHQUFUVEFHQUdHQUNBQVRBVFRBQVRBQVRBQ0NBVENHQ0FDQUdUR1RUVENUVFRHVFRHVFRBQVRHQ1RBQ0FBQ0FUQUNBQUFHQUdHQUFHQ0FUR0NBRyIKbXlfZG5hCmxlbmd0aChteV9kbmEpCmNsYXNzKG15X2RuYSkKc3RyKG15X2RuYSkKbmNoYXIobXlfZG5hKQpgYGAKCmBgYHtyIGRuYV9leDMsIGluY2x1ZGUgPSBULCBlY2hvPVQsIGV2YWw9VH0KbXlfZG5hX2xpc3QgPC0gc3Ryc3BsaXQoeCA9IG15X2RuYSwgc3BsaXQgPSAiIiwgZml4ZWQgPSBUUlVFKQpsZW5ndGgobXlfZG5hX2xpc3QpCmNsYXNzKG15X2RuYV9saXN0KQpteV9kbmFfdmVjdG9yIDwtIHVubGlzdChteV9kbmFfbGlzdCkKbGVuZ3RoKG15X2RuYV9saXN0W1sxXV0pCnN0cihteV9kbmFfdmVjdG9yKQpsZW5ndGgobXlfZG5hX3ZlY3RvcikKCiMgdW5pcXVlIGNoYXJhY3RlcnMKdW5pcXVlKG15X2RuYV92ZWN0b3IpCgojIG51bWJlciBvZiBBcwoobXlfZG5hX3ZlY3RvciA9PSAiQSIpCmxlbmd0aChteV9kbmFfdmVjdG9yW215X2RuYV92ZWN0b3IgPT0gIkEiXSkKCmBgYAoKCiMjIEZyZXF1ZW5jeSBkaXN0cmlidXRpb24KQSBnZW5lIGNvbnNpc3RzIG9mIGEgc2VxdWVuY2Ugb2YgbnVjbGVvdGlkZXMge0EsIEMsIEcsIFR9LlRoZSBudW1iZXIgb2YgZWFjaCBudWNsZW90aWRlIGNhbiBiZSBkaXNwbGF5ZWQgaW4gYSBmcmVxdWVuY3kgdGFibGUuIFRoaXMgd2lsbCBiZSBpbGx1c3RyYXRlZCBieSB0aGUgWnl4aW4gZ2VuZSB3aGljaCBwbGF5cyBhbiBpbXBvcnRhbnQgcm9sZSBpbiBjZWxsIGFkaGVzaW9uIChHb2x1YiBldCBhbC4sIDE5OTkpLiAgVGhlIGFjY2Vzc2lvbiBudW1iZXIgKFg5NDk5MS4xKSBvZiBvbmVvZiBpdHMgdmFyaWFudHMgY2FuIGJlIGZvdW5kIHZpYSBhbiBOQ0JJIFVuaUdlbmUgc2VhcmNoLiBUaGUgY29kZSBiZWxvdyBpbGx1c3RyYXRlcyBob3cgdG8gaW5zdGFsbCB0aGUgcGFja2FnZSBhcGUsIHRvIGxvYWQgaXQsIHRvIHJlYWQgZ2VuZSDigJ1YOTQ5OTEuMeKAnW9mIHRoZSBzcGVjaWVzIGhvbW8gc2FwaWVucyBmcm9tIEdlbkJhbmssIGFuZCB0byBtYWtlIGEgZnJlcXVlbmN5IHRhYmxlIG9mIHRoZSBmb3VyIG51Y2xlb3RpZGVzCgpgYGB7ciBmcmVxMSwgaW5jbHVkZSA9IFQsIGVjaG89VCwgZXZhbD1UfQojIGluc3RhbGwucGFja2FnZXMoYygiYXBlIiksIHJlcG89Imh0dHA6Ly9jcmFuLnItcHJvamVjdC5vcmciLCBkZXA9VFJVRSkKbGlicmFyeShhcGUpCmdlbmUgPC0gcmVhZC5HZW5CYW5rKGMoIlg5NDk5MS4xIiksICBhcy5jaGFyYWN0ZXI9VFJVRSkKdGFibGUoZ2VuZSkKcGllKHRhYmxlKGdlbmUpKQpgYGAKCmBgYHtyIGtuaXRfZXhpdCwgaW5jbHVkZT1GLCBlY2hvPUZ9CmtuaXRfZXhpdCgpCmBgYAo=